Overview of the Ubiquitin-Proteasome System for Protein Degradation
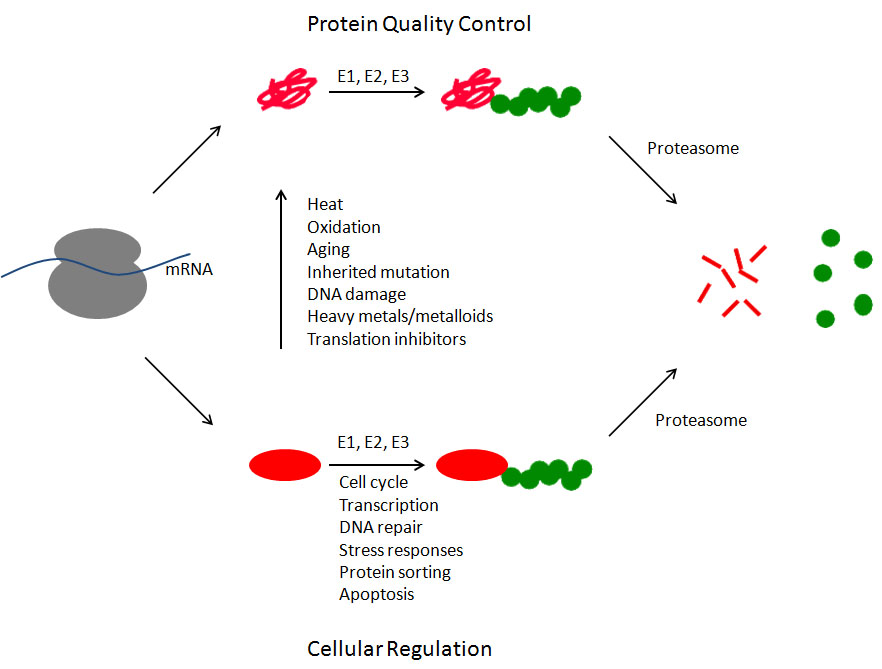 Many proteins misfold either during or immediately after protein synthesis (upper pathway). These proteins (shown in red) are recognized by a series of ubiquitinating enzymes (known as E1, E2, and E3) that covalently attach the small protein ubiquitin (shown in green) to the misfolded protein. This ubiquitin signal allows the protein to be recognized by the proteasome. This 2.5 mDa molecular machine binds the ubiquitinated substrate, unfolds the substrate, removes the ubiquitin molecules, and injects the polypetide chain into the central core of the proteasome where the protein is degraded into small polypeptides. Properly folded proteins are also subject to degradation (lower pathway). In these cases, by controlling the abundance of key regulatory proteins, the ubiquitin-proteasome system contributes to nearly every aspect of cell biology. Some of the best characterized examples are listed. And, of course, any properly folded protein can be converted into a misfolded protein at any time due to exposure to any of the numerous stressors that cause protein misfolding, only a few of which are listed.
Many proteins misfold either during or immediately after protein synthesis (upper pathway). These proteins (shown in red) are recognized by a series of ubiquitinating enzymes (known as E1, E2, and E3) that covalently attach the small protein ubiquitin (shown in green) to the misfolded protein. This ubiquitin signal allows the protein to be recognized by the proteasome. This 2.5 mDa molecular machine binds the ubiquitinated substrate, unfolds the substrate, removes the ubiquitin molecules, and injects the polypetide chain into the central core of the proteasome where the protein is degraded into small polypeptides. Properly folded proteins are also subject to degradation (lower pathway). In these cases, by controlling the abundance of key regulatory proteins, the ubiquitin-proteasome system contributes to nearly every aspect of cell biology. Some of the best characterized examples are listed. And, of course, any properly folded protein can be converted into a misfolded protein at any time due to exposure to any of the numerous stressors that cause protein misfolding, only a few of which are listed.
Areas of Study
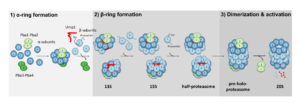
1. Structural analysis of proteasome core particle assembly. The proteasome consists of a core particle and a regulatory particle. Proteolysis occurs within the central barrel-shaped CP, while access of substrates to the CP is mediated by the RP. The CP consists of four stacked rings and is a 700 kDa 28-subunit complex. It cannot assembly spontaneously, but rather is built through an ordered multistep pathway that requires 5 dedicated chaperones: Pba1/2, Pba3/4, and Ump1 (see figure). We have developed methods to obtain high resolution cryo-EM structures of these normal low abundance transitory intermediates for the first time. This work is being conducted in collaboration with the Harvard Cryo-EM Center for Structural Biology.
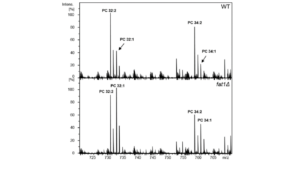
2. Role of lipid metabolism in protein quality control. Increasing evidence suggests a very close relationship between pathways of lipid and protein homeostasis. The unfolded protein response (UPR) monitors and responds to the presence of misfolded proteins in the endoplasmic reticulum. Recent evidence indicates that the UPR can also be induced by various defects in lipid metabolism. Conversely, some of the UPR’s transcriptional targets include genes of lipid metabolism. We have developed powerful lipidomics approaches, in collaboration with the lab of Dr. Nathalie Agar, to study the relationship between lipid metabolism and protein quality control.
Comments: 0